Understanding the complexity of cancer requires methods that can integrate equally complex biological data. In our lab, we are committed to developing probabilistic models that bring together multiple molecular layers, including genomics, epigenomics, transcriptomics, proteomics and metabolomics, to provide a holistic view of each patient. These models uncover hidden structure by capturing both shared and modality-specific variation, allowing us to reduce noise and reveal biologically meaningful patterns. By modeling system-level responses to perturbations such as drug treatments or environmental changes, we aim to generate representations that are not only statistically robust but also interpretable, enabling new biological insights that can be directly validated and translated into clinical understanding.
MuVI
MuVI is a general-purpose probabilistic latent variable model for multi-omics integration that incorporates prior biological knowledge into its structure. It uses pathway annotations, gene sets, or cell-type signatures to guide the discovery of latent factors that explain variation across different data types. Even when this prior knowledge is noisy or incomplete, MuVI is able to learn biologically relevant dimensions, enabling scientists to interpret the sources of variation in the data more clearly and to relate them to known mechanisms.

MUSIC
MUSIC (MUltiview baySIan tensor deComposition) extends probabilistic modeling to high-dimensional array data, such as time-series or condition-specific measurements. It jointly decomposes collections of heterogeneous tensors, e.g. patient × gene × time or patient × protein × condition, into shared and modality-specific components. With structured sparsity priors and efficient variational inference, MUSIC scales to large datasets, handles missing data, and yields interpretable embeddings. We have applied it to cancer drug-response studies and single-cell leukemia data, where it revealed meaningful molecular signatures associated with disease pathways.
MOMO-GP
MOMO-GP (Multi-Omic Multi-output Gaussian Processes) addresses the challenge of learning interpretable representations from single-cell multi-omics data, which are typically high-dimensional, sparse, and nonlinear. Unlike traditional methods that trade off interpretability for modeling power, MOMO-GP combines neural networks with Gaussian Processes to achieve both. It learns separate latent embeddings for cells and features, as well as shared and modality-specific components in the multi-view setting. By modeling gene relevance explicitly, MOMO-GP connects cell clusters to marker genes, making the learned structure readily interpretable in biological terms.
JOANA
JOANA is a probabilistic model for pathway enrichment analysis (PEA) that overcomes limitations of classical approaches like Over-Representation Analysis (ORA) and Functional Class Scoring (FCS). While methods such as GSEA work with continuous scores, they typically operate on a single omics layer and can yield overly broad sets of enriched pathways. JOANA improves on this by modeling enrichment scores across multiple omics layers using mixtures of beta distributions within a Bayesian framework. This allows it to estimate the probability of pathway enrichment both within and across modalities, yielding higher precision and more biologically relevant results.
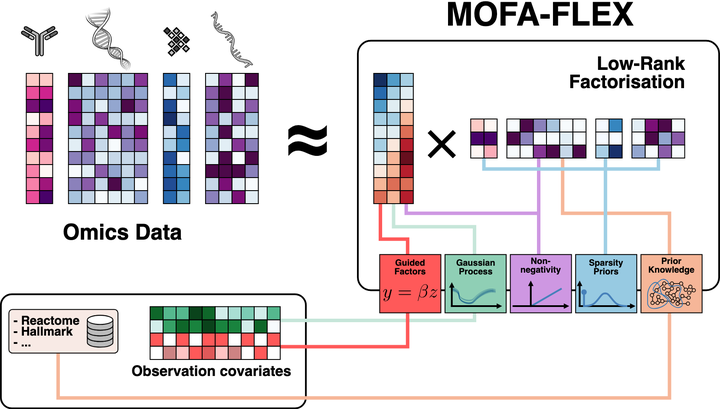
MOFAFLEX
MOFAFLEX is our upcoming framework for flexible and interpretable multi-omics integration. Designed to generalize the principles behind models like MuVI and MUSIC, MOFAFLEX supports heterogeneous data types, modular priors, and scalable inference. Its architecture allows for tailored modeling of real-world datasets, balancing interpretability with modeling flexibility. MOFAFLEX is currently under active development and will provide a unified foundation for future applications in cancer biology and beyond.
